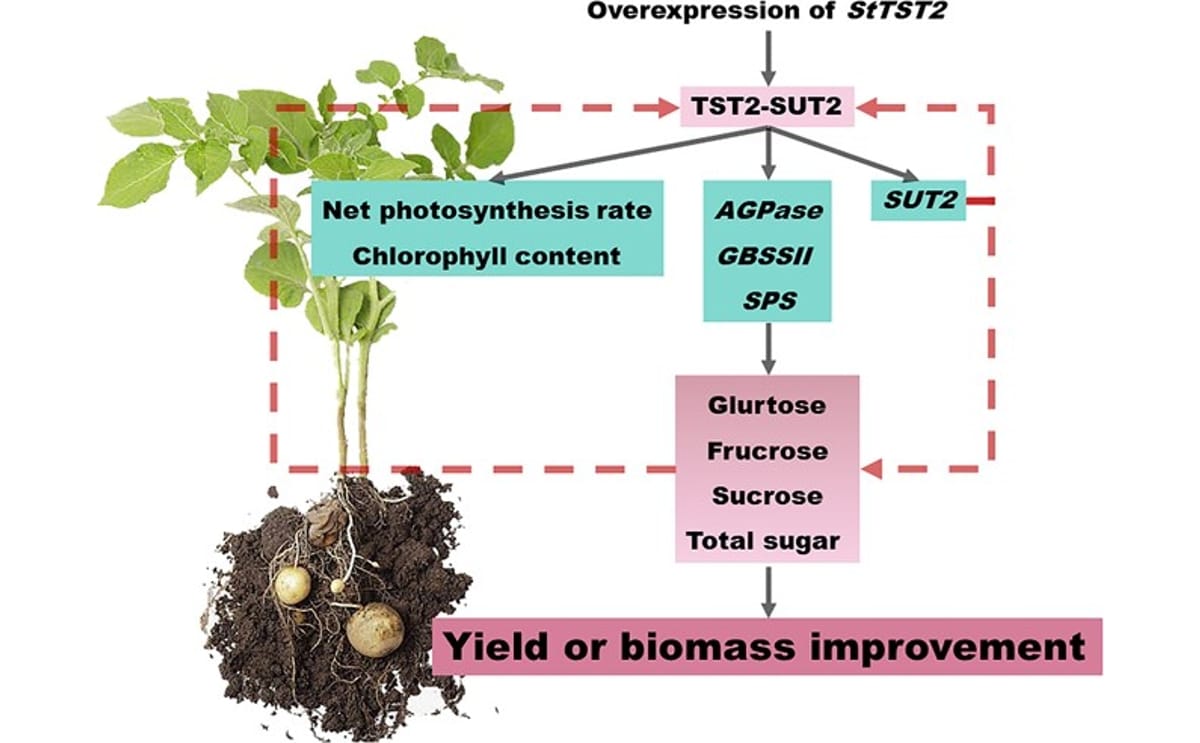
StTST2–SUT2 regulatory module in potato. Confocal and protein interaction analyses show StTST2 on the vacuolar membrane interacting with SUT2, enhancing sugar transport from leaves to tubers, boosting photosynthesis and yield, and highlighting its key role in sugar accumulation and tuber productivity.
Haplotype-Resolved Potato Genome Reveals Key Gene Driving Sugar Accumulation and Yield
Scientists have achieved a landmark breakthrough in potato genomics by generating a chromosome-scale, haplotype-resolved genome and uncovering a key gene that directly enhances sugar content, photosynthesis, and tuber yield. The study, published in Horticulture Research (2025), provides one of the most comprehensive genomic resources ever developed for cultivated potato and offers powerful new tools for precision breeding.
Potato (Solanum tuberosum L.) is the fourth most important food crop worldwide, yet its genetic improvement has lagged behind cereals due to its autotetraploid genome, extreme heterozygosity, and complex inheritance patterns. This research addresses those challenges head-on.
Phenotypes, chromosomes, and genome assembly of ‘N8–4’

Phenotypes and genomic assembly of ‘N8–4’. (a) Anther-cultured heterozygous diploid ‘N8–4’ seedlings from tetraploid ‘Ningshu 15’. Scale bar = 2 cm. (b) Representative tubers of ‘N8–4’. Scale bar = 2 cm. (c, d) Fluorescence in situ hybridization (FISH) showing chromosome patterns of ‘N8–4’ and ‘N15’. Scale bar = 5 μm. (e) Hi-C interaction heatmap of 12 chromosomes in two haplotypes of ‘N8–4’; dark red indicates high contact probability. (f) Genomic features of the two haplotypes: chromosome length, gene density, transposable elements, GC content, and gene expression levels, with syntenic relationships indicated.
To overcome polyploid complexity, researchers used ‘N8–4’, a diploid anther-cultured progeny derived from the elite tetraploid cultivar ‘Ningshu 15’, known for high starch content and drought resistance. Although ‘N8–4’ showed dwarf growth compared with its parent, it retained stable starch characteristics, making it ideal for genome assembly.
By integrating PacBio long-read sequencing, Illumina short-read sequencing, and Hi-C chromatin interaction data, the team assembled a 1.653 Gb genome, fully resolved into two haplotypes:
- Haplotype 1: 828.21 Mb
- Haplotype 2: 825.83 Mb
Quality indicators confirm a world-class assembly:
- Contig N50: ~1.47–1.48 Mb
- Scaffold N50: 61–65 Mb
- BUSCO completeness: 93.6–94.2%
- LTR Assembly Index (LAI): 11.62 and 11.94
More than 94% of sequences were anchored to 12 chromosomes per haplotype, and ~58% of the genome consists of transposable elements, dominated by long terminal repeats. Each haplotype contains ~34,000–35,000 protein-coding genes, with extensive functional annotation across GO, KEGG, Pfam, InterPro, Swiss-Prot, and TrEMBL databases.
Comparative genomics and evolutionary signatures

GWAS of tuber glucose content and population structure. (a) Phenotypic diversity of tubers in 87 potato accessions and their phylogenetic tree. (b) STRUCTURE analysis showing five subgroups in the GWAS population. (c) PCA of 141 accessions based on SNPs; clusters correspond to genetic similarity. (d) Manhattan plot identifying a significant QTL for glucose content on chromosome 5; regional plot shows a 185 kb candidate interval.
Comparative analyses with tomato, pepper, tobacco, rice, sweet potato, and reference potato genomes revealed:
- Over 10,800 shared orthogroups, including 239 single-copy genes
- Positive selection in genes involved in:
- Starch and sucrose metabolism
- Fructose and mannose metabolism
- Amino sugar and nucleotide sugar pathways
- Carotenoid biosynthesis
Gene family expansion was strongly enriched in sugar metabolism, ribosome function, and protein processing, while contraction occurred in photosynthesis- and hormone-related pathways. Large-scale inversions, translocations, and duplications were detected between haplotypes and the DM reference genome, highlighting structural variation as a driver of agronomic diversity.
GWAS population structure and tuber diversity

Overexpression of StTST2 in potato enhances growth, sugar accumulation, and yield. (a) 90-day-old T0 transgenic plants; (b) tuber phenotypes of WT and T0 overexpression (OE) plants; (c) 60-day-old T1 transgenic plants; (d,e) yield of WT and OE T0 and T1 plants; (f) net photosynthetic rate; (g) leaf chlorophyll content. Transgenic lines show increased tuber size, yield, photosynthesis, and sugar accumulation compared with WT (mean ± SE, n = 3; *P < 0.05, **P < 0.01).
To connect genomic variation with agronomic traits, the researchers resequenced 141 tetraploid potato accessions including modern cultivars, historical varieties, CIP lines, and breeding materials. Each accession was sequenced to ~10× depth, generating ~18 million high-quality SNPs.
Population structure analysis (neighbor-joining tree, PCA, STRUCTURE) divided accessions into five genetic clusters. These clusters showed clear differences in tuber glucose levels, confirming that sugar content is shaped by genetic background and breeding history. Notably, drought-resistant varieties such as Ningshu 15 and Qingshu 9 clustered together, validating the biological relevance of the population structure.
Genome-wide association study identifies sugar loci
Using GLM, MLM, and FarmCPU models, the GWAS identified 53 robust quantitative trait loci (QTLs) associated with tuber sugar content:
- 38 loci for glucose
- 11 loci for sucrose
- 4 loci for fructose
These loci were distributed across multiple chromosomes, confirming that sugar content is a highly polygenic trait. A single, strong association peak on chromosome 5 stood out across models, explaining a significant portion of glucose variation. This region led to the identification of StTST2, a gene encoding a tonoplast sugar transporter.
Functional validation of StTST2 in potato and Arabidopsis
To validate function, StTST2 was overexpressed in potato cultivar ‘Atlantic’:
- Larger tubers and significantly higher yield
- Increased leaf and tuber glucose, fructose, sucrose, and total sugars
- Higher photosynthetic rate and chlorophyll content
When expressed in Arabidopsis, StTST2 similarly enhanced biomass accumulation, leaf sugar levels, and seed yield, demonstrating a conserved role across species.
Molecular mechanism: StTST2–SUT2 regulatory module
Subcellular localization experiments showed StTST2 resides on the vacuolar membrane (tonoplast). Protein–protein interaction assays confirmed that StTST2 physically interacts with SUT2, a sucrose transporter and sensor.
This interaction:
- Upregulates key metabolic genes (AGPase, GBSSII, SPS, SUT2)
- Enhances intracellular sugar partitioning
- Strengthens source-to-sink transport from leaves to tubers
- Creates a positive feedback loop linking sugar sensing, photosynthesis, and yield formation
Why this discovery matters
This study delivers three major advances:
- A high-quality haplotype-resolved potato genome: suitable for allele-specific expression and structural variation studies
- 53 validated sugar-related QTLs: for marker-assisted and genomic selection
- A functionally confirmed yield-enhancing gene (StTST2): offering a direct target for breeding and biotechnology
Together, these findings significantly advance our understanding of sugar metabolism, yield regulation, and genome evolution in potato, and provide a strong foundation for developing high-yielding, high-quality, and stress-resilient potato varieties to support future global food security.
The findings are reported in the journal Horticulture Research (2025) and can be accessed at:https://doi.org/10.1093/hr/uhaf075